Day 1 :
Keynote Forum
Prof. Martin Ryan
University of St Andrews, UK
Keynote: Protein biogenesis in the Picornaviridae : Virus-encoded proteinases, ribosome ‘skipping’, alternative initiation and programmed ribosomal frame-shifting
Time : 09:00-09:40

Biography:
Martin D Ryan has cloned and sequenced the genome of Enterovirus Type 70 as a part of his Ph.D. research work. He then went on to work on FMDV polyprotein processing at the Pirbright Institute. After moving to University of St. Andrews in 1994 he continued this work, focusing on the mechanism of FMDV 2A-mediated ‘cleavage’ – showing it was not a proteolytic element, but mediated a novel form of translational recoding – ‘ribosome skipping’. Other lines of research led to the development of 2A as a protein co-expression system – now used very widely in biotechnology and biomedicine. In 2017, he was awarded the Unilever Colworth Prize for his work on 2A. More recently, his laboratory has been working on FMDV RNA replication using a replicon system encoding fluorescent proteins such that FMDV RNA replication can be monitored by live-cell imaging.
Abstract:
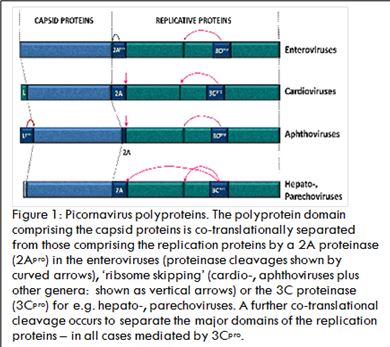
The genome structure of viruses within the Picornavirus family is similar to cellular mRNAs: +v sense RNA genomes comprising a long 5’ non-coding region (NCR), a single open reading frame (ORF; ~2,300 aa), a short 3’-NCR with a poly(A) tail, although the 5’ cap structure is quite different. When introduced into the cytoplasm, the virus RNA can function directly as a messenger RNA. The challenge faced by these viruses is, however, the need to generate a multiplicity of different proteins (capsid, RNA replication proteins) from this single ORF. All picornaviruses encode a proteinase (3Cpro) which ‘processes’ the polyprotein into mature products by a combination of a single, co-translational, intramolecular cleavage (in cis) and multiple post-translational intermolecular cleavages (in trans). Some picornaviruses have acquired a second proteinase (aphthoviruses – Lpro; enteroviruses – 2Apro) which perform a single co-translational cleavage in cis, but then go onto cleave cellular proteins involved in the cap-dependent translation of host-cell mRNAs and proteins involved in interferon production in response to infection. The 2A region of aphtho- and many other Picornavirus genera (unlike enteroviruses not a proteinase) may either be a short oligopeptide sequence, or, a larger protein that mediates a translational ‘recoding event termed ‘ribosome skipping’ at the C-terminus of 2A such that translation stops (releasing the nascent protein) then recommences translation of the down-stream replication protein sequences: an apparent ‘cleavage’, but in actuality a discontinuity in the polypeptide backbone. It is now known that this form of control over protein biogenesis is used by other types of RNA virus and some cellular genes. Uniquely within the picornaviruses, Theiler's murine encephalomyelitis virus (TMEV; genus Cardiovirus) encodes an overlapping, alternative ORF within the leader, or L protein, alternative initiation producing the L* protein which plays an important role in the establishment of persistent CNS infections by TMEV. Recently it has been shown that some picornaviruses also use programmed ribosomal frame-shifting shortly after the ribosome skipping event to (translationally) down-regulate the production of replication proteins at later stages of the infectious cycle. Furthermore, this frame-shifting does not rely solely on an RNA secondary structure but is directed by protein 2A. In conclusion, the picornaviruses have evolved a range of different mechanisms to control their protein biogenesis: not at the level of RNA transcription, but by co- and post-translational events alone.
Recent Publications
- Ryan MD, Flint M (1997) Virus-encoded proteinases of the picornavirus super-group. J. Gen. Virol. 78: 699-723.
- Donnelly MLL, Luke G, Mehrotra A, Li X, Hughes LE, Gani D, Ryan MD (2001) Analysis of the aphthovirus 2A/2B polyprotein ‘cleavage’ mechanism indicates not a proteolytic reaction, but a novel translational effect: a putative ribosomal ‘skip’. J. Gen. Virol. 82:1013-1025.
- Sharma P, Yan F, Doronina V, Escuin-Ordinas H, Ryan MD, Brown J (2012) 2A peptides provide distinct solutions to driving stop-carry on translational recoding. Nuc. Acids Res. 40:1-9.
- Chen HH, Kong WP, Zhang L, Ward PL, Roos RP (1995) A picornaviral protein synthesized out of frame with the polyprotein plays a key role in a virus-induced immune-mediated demyelinating disease. Nat. Med. 1:927-931.
- Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE (2017) Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 8:15582.
Keynote Forum
Maurizio Provenzano
University Hospital of Zürich, Switzerland
Keynote: The role of the human polyomavirus BK in the development of prostate cancer
Time : 09:40-10:20

Biography:
Maurizio Provenzano graduated as a MD from La Sapienza University of Rome and obtained his specialization in Medical Oncology at the University of Milan and his Ph.D. in Virology at the University of Padua. He is a tenured investigator in the position of the Head of Research at the Department of Urology, University Hospital of Zurich and Lecturer at the Faculty of Medicine, University of Zurich. Since 2007, he has been a member of the Cancer Network of Zurich. He has authored more than 70 peer-reviewed publications including, reviews, book chapters, and conference papers, many of which are in highly qualified scientific periodicals and has been serving as an Editorial Board Member of reputed journals. He has been the recipient of professional awards and prizes for the contribution to the field of Viral Oncology and Onco-Immunology.
Abstract:
In recent years the scientific literature in the field of the prostate carcinoma (PCa) pointed out on the genetic heterogeneity occurring in this tumour, while little attention was given to the causes that threaten the genomic integrity of the prostate. Together with chemical and physical agents, biological agents such as viruses with oncogenic potential, which interfere with the cell cycle, are responsible for gene alterations and might be included in the putative genomic evolution of PCa. Polyomavirus BK (BKPyV) encodes two viral oncogenes, the large T antigen (LTag) and small t antigen (tag). Transformation of animal and human cells by BKPyV is operated by these two viral oncoproteins. LTag binds and abolishes the functions of the tumour suppressor p53 and pRB family proteins, whereas tag interacts with the phosphatase PP2A, which activates the Wnt pathway. Moreover, tag activates the phosphatidylinositol 3-kinase, an enzyme involved in pathways crucial for cell proliferation and transformation. LTag is also clastogenic and mutagenic. These activities are able to hit the cellular genome, which accumulates many gene mutations/chromosome aberrations. Considering that BKPyV infection is ubiquitous in the general population, it is difficult to assess a specific role of this virus in cellular transformation. BKPyV is kidney-tropic and remains latent in many human organs/tissues, mainly the urogenital tract. The lifelong period of BKPyV infection, together with the fact that about 95% of PCa are slow-growing organ-confined indolent tumors, could be responsible of those gene/chromosomal damages, which initiate the development of this malignancy. Footprints of this small DNA tumour virus has been revealed at higher prevalence in early stages PCa than in healthy control tissues, thus providing an indication for an increased risk of PCa development with the presence of BKPyV infection. Loss-of-function mutations in p53 gene at very early stages of PCa are rare. Therefore, the sequestration of wild-type p53 exerted by BKPyV LTag oncoprotein at initial phases is a hallmark for BKPyV involvement and may ensure the genetic heterogeneity of PCa.
Keynote Forum
Joachim Wink
Helmholtz Centre for Infection Research, Germany
Keynote: Polyphasic taxonomy – The Class Actinobacteria
Time : 11:00-11:40

Biography:
Joachim Wink has completed his PhD in 1985 from Frankfurt University. He then went to the pharmaceutical industry and started his career at the Hoechst AG, where he was responsible for the strain collection and specialized in the cultivation and taxonomic characterization of Actinobacteria and Myxobacteria. During the years, he was responsible for the strain library within the pharmaceutical research and a number of screening projects with Hoechst Marion Russel, Aventis and Sanofi. In the year 2005, he did his habilitation at the Carolo Wilhelma University of Braunschweig and in 2012 he went to the Helmholtz Centre for Infection Research in Braunschweig, where he founded the working group of the strain collection with its focus on Myxobacteria. Here, he is now working on the isolation and taxonomic characterization of Myxobacteria and Actinobacteria, as well as, the analysis of their secondary metabolites. He has published more than 50 papers on secondary metabolites and the taxonomy of the producing microorganisms in reputed journals, a number of reviews, as well as, book chapters and more than 35 patents. He is the Editorial Board Member of a number of international journals.
Abstract:
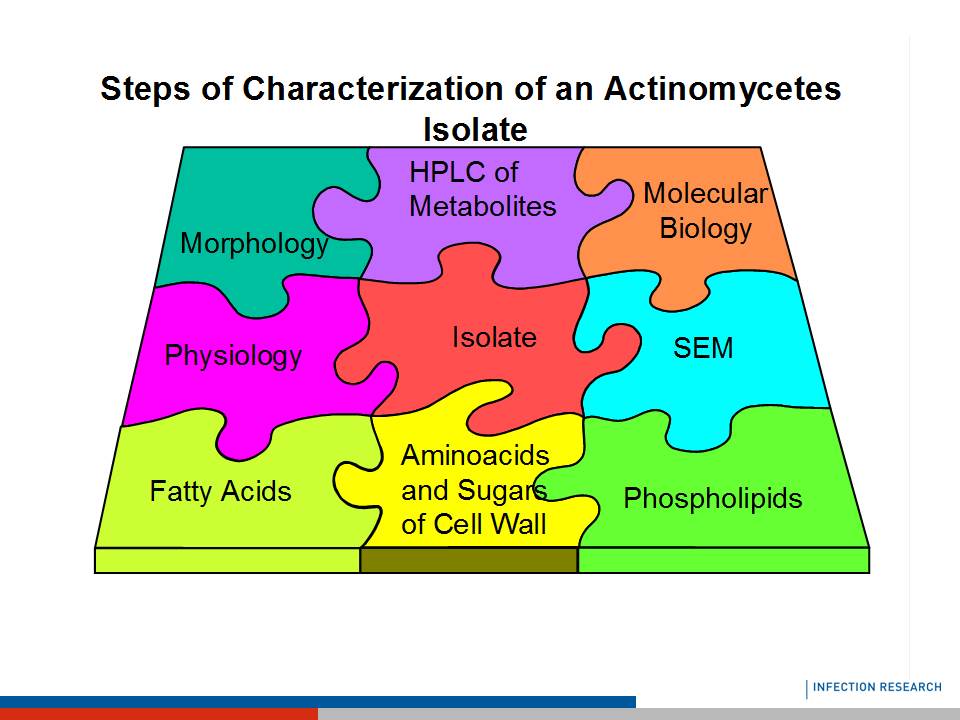
Members of the class Actinobacteria plays an important role for humans, on one hand, we have producer of antibiotics, which mainly belong to the so called Sporoactinomycetes like Streptomyces or Amycolatopsis and on the other hand the pathogenic ones like many species of the Mycobacteria and Nocardia. Actinobacteria are typical soil bacteria, but also inhabit many other habitats, so that they could be isolated from different materials. Even their morphology has been used for the characterization for many years; today a number of molecular biological methods are also available for their taxonomic classification. In the 90th of the last century the polyphasic approach for the classification of bacteria was introduced and is also today the way to identify a species of the Actinobacteria. This approach combines the methods of the morphology and physiology, together with the chemotaxonomic methods and the methods of the molecular biological characterization. The wide variety of the morphology of the different Actinobacteria, we find coccoid and rod shaped single cell forming species like in the Micrococcales, as well as, organisms that form mycelia and differentiate with spore chains or sporangia, made it very important for the identification and classification of this bacteria to have a number of methods available. Especially after finding the different morphological features also in different genera and families the chemotaxonomy became very important in the 60th of the last century. The molecular biological characterization methods opened a number of additional tools, but there are many examples that they also cannot stand alone. Aim of the workshop is after a short introduction of the bacterial class to show the different aspects of the polyphasic taxonomy basing on the work of Vandamme et al. with the class Actinobacteria as an example. It will be discussed if the molecular biological characterization will replace morphology and chemotaxonomy.
Recent Publications
- Landwehr W, Wolf C, Wink J (2016) Actinobacteria and Myxobacteria – Two of the Most Important Bacterial Resources for Novel Antibiotics. in M. Stadler and P. Dersch (eds.) How to Overcome the Antibiotic Crisis. Springer. Volume 398 of the series Current Topics in Microbiology and Immunology. 273-302.
- Wink J, Schumann P, Klenk HP, Zaburannyi N, Westermann M, Martin K, Glaeser SP, Kämpfer P, Atasayar E (2016) Streptomyces “caelicus" an antibiotic producing species of the genus Streptomyces and Streptomyces canchipurensis Li et al. 2014 are later heterotypic synonym of Streptomyces muensis Ningthoujam et al. 2013. Int J Syst Evol Microbiol. doi: 10.1099/ijsem.0.001612. [Epub ahead of print].
- Mohammadipanah, J Wink (2016) Actinobacteria from Arid and Desert Habitats: Diversity and Biological Activity. Front Microbiol 2015 6:1541. doi: 10.3389/fmicb.2015.01541.
- Wink J, Schumann P, Spöer C, Eisenbarth K, Glaeser SP, Martin K, Kämpfer P (2014) Emended descriptions of Actinoplanes friuliensis and description of Actinoplanes nipponensis sp. nov., two Antibiotic Producing Species of the Genus Actinoplanes. IJSEM 64:599-606.
- Müller R, Wink J (2014) Future potential for anti-infectives from bacteria – How to exploit biodiversity and genomic potential. IJMM 304:3-13.5.
- Bacteriology, Virology, Veterinary Microbiology, Mycology, Phycology, Environmental and Agricultural Microbiology
Location: Redwood Suite C

Chair
Joachim Wink
Helmholtz Centre for Infection Research, Germany

Co-Chair
Stef Stienstra
Scientific Advisor Royal Dutch Navy, Netherlands
Session Introduction
Maria Angela Cunha
University of Aveiro, Portugal
Title: Photodynamic inactivation of phytopathogenic fungi with natural and synthetic photosensitizers
Time : 11:40-12:05

Biography:
Angela Cunha is an Assistant Professor in the Biology department of the University of Aveiro, Portugal and a Researcher at the Centre for Environmental and Marine Studies. She has been involved in lecturing graduate and post graduate courses in the fields of Microbiology and Genetics and is currently the Director of the Bachelor Course in Biology. Her main research field is the distribution and activity of microorganisms in the environment, microbial bioremediation tools and interactions of microorganisms with plants in the perspectives of disease control, plant growth promotion and microbe-assisted phytoremediation.
Abstract:
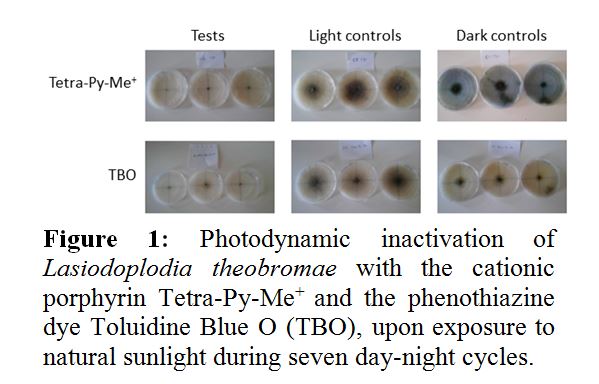
Diseases caused by phytopathogenic fungi on vine and fruit trees are associated with important economic losses. With the aim of reducing the environmental impacts of traditional fungicides used in agriculture, innovative approaches to inactivate fungi in environmental matrices are being investigated. In this context, the photodynamic effect, in which the combination of a photosensitizer, visible light and molecular oxygen leads to the formation of cytotoxic species of oxygen, represents a promising perspective. The objective of this work was to evaluate the efficiency of photodynamic inactivation (PDI) of model phytopathogenic fungi (Lasiodoplodia theobromae and Botrytis cinerea) with natural (curcumin and riboflavin) and synthetic (cationic porphyrin Tetra-Py-Me+ and toluidine blue O) photosensitizers. Exposure to natural sunlight during 7 day-night cycles, in presence of 500 µM of Tetra-Py-Me+ or 500 µM toluidine blue O, attenuated the growth of L. theobromae by 35% and 26%, respectively (Figure 1). Curcumin and riboflavin failed to cause significant inactivation. Light alone (light control) and exposure to the photosensitizer in the absence of light (dark control) did not affect the growth of L. theobromae. On the contrary, the growth of B. cinerea was significantly inhibited by light alone. Tetra-Py-Me+ and toluidine blue O caused 90% and 93% attenuation of growth, respectively, in relation to the dark control. PDI with the natural photosensitizers was also not significant. The results confirm that PDI with porphyrins or phenothiazines may be further explored as a viable alternative to chemical fungicides in the control of phytopathogenic fungi. However, repeated treatments may be required considering that in field conditions, fungi may recover from sub-lethal damage, during the night period.
Recent Publications
- Fernández L, Esteves VI, Cunha A, Schneider RJ, Tomé JPC (2016) Photodegradation of organic pollutants in water by immobilized porphyrins and phthalocyanines. Journal of Porphyrins and Phthalocyanines 20:150–166.
- Rocha DM, Venkatramaiah N, Gomes MC, Almeida A, Faustino, MA, Paz FA, Cunha Â, Tomé JP (2015) Photodynamic inactivation of Escherichia coli with cationic ammonium Zn (ii) phthalocyanines. Photochemical & Photobiological Sciences 14(10):1872-1879.
- Beirão S, Fernandes S, Coelho J, Faustino MAF, Tomé JPC, Neves MGPMS, Tomé A, Almeida A, Cunha A (2014) Photodynamic inactivation of bacterial and yeast biofilms with a cationic porphyrin. Photochemistry & Photobiology 90(6):1387–1396.
- Gomes MC, Silva S, Faustino MAF, Neves MGPMS, Almeida A, Cavaleiro, JAS, Tomé JPC, Cunha A (2013) Cationic galactoporphyrin photosensitisers against UV-B resistant bacteria: oxidation of lipids and proteins by 1O2. Photochemical & Photobiological Sciences 12:262–271.
- Gomes MC, Woranovicz-Barreira SM, Faustino MAF, Fernandes R, Neves MGPMS, Tomé AC, Gomes NCM, Almeida A, Cavaleiro, JAS, Cunha A, Tomé JPC (2011) Photodynamic inactivation of Penicillium chrysogenum conidia by cationic porphyrins. Photochemical & Photobiological Sciences 10:1735-1743.
Rehana A.Gilani
Shaheed Zulfiqar Ali Bhutto Medical University, Pakistan
Title: Resistance profile of ESBL positive uropathogenic E. coli in a tertiary care teaching hospital in Islamabad, Pakistan

Biography:
Rehana A Gilani has completed her MPhil from a teaching hospital in Pakistan and has tried to incorporate research in the regular curriculum of the newly established university.
Abstract:
Introduction & Aim: Escherichia coli is the most common Gram negative organism causing both community, as well a,s hospital acquired Urinary Tract Infection (UTI). This study was conducted to find out the prevalence of uropathogenic E. coli and to demonstrate ESBL phenomenon and their antimicrobial susceptibility patterns. The genetic characterization of ESBL producing isolates was also done to demonstrate the genes conferring drug resistance to these organisms in our institution using conventional PCR.
Materials & Methods: This research was a prospective, non- randomized, descriptive study. A pro forma was used as a tool for data collection. The study was conducted in the Microbiology Laboratory, Department of Pathology, PIMS, Shaheed Zulfiqar Ali Bhutto Medical University for 8 months. 140 urine non-duplicate samples of patients with UTI yielding growth of E. coli were selected and their susceptibility profile was determined.
Results: Urine samples of 140 patients yielding E. coli were enrolled in the study. There were 81 (58%) females and 59 (42%) males. Patients included in the study were from 12-86 years of age. The ESBL phenomenon was confirmed by double disc method, which demonstrated that 80 (57%) samples were positive and 60 (43%) were negative.
Conclusion: Our results showed emergence of multidrug resistant ESBL producing E. coli in our set up, which is a very serious problem due to non-availability of new antibiotics and also the element of poverty, due to which the patients are unable to afford the prescribed drugs.
Herbert B. Allen
Drexel University College of Medicine, USA
Title: The Pathway to Alzheimer’s Disease

Biography:
Herbert B Allen specialties include dermatology and dermatopathology, skin pathology and fungal infections. He is a graduate of Johns Hopkins University School of Medicine. He has served on the boards of the American Society of Dermatology and the American College of Physicians and has published over 30 scientific articles in the fields of dermatology and dermatopathology. He is the author of ‘Keywords’ in Dermatology, a book on the language of dermatology. He's board-certified with the American Board of Dermatology and the American Board of Pathology. He is currently an Emeritus Professor in the department of Dermatology where he served as chair of the department for 14 years.
Abstract:
The pathway to Alzheimer’s disease (AD) follows a route which many chronic diseases utilize: microbes make biofilms that activate the innate immune system which ultimately leads to tissue destruction. The primary microbes involved are very likely dental and Lyme spirochetes. Both have been found by PCR; additionally, Lyme spirochetes have been cultured from affected brains. Both are known to make biofilms, and these biofilms have receptor sites for Toll-like receptor 2 (TLR2). In its activity as a first responder, TLR2 makes use of the myeloid differentiation 88 pathway to kill invading microorganisms. This pathway generates NFκB and TNFα and these molecules in turn appear not only to be responsible for destruction of neurons, but also to be responsible, in large part, for the creation of beta amyloid. (Aβ) The Aβ has been previously thought to be primary in the AD pathogenesis, but it turns out to be antimicrobial. Neither the TLR2 nor the Aβ can penetrate the biofilms and thus they destroy the surrounding tissue as “innocent bystanders”. The recent findings of intracellular, in contrast to extracellular, biofilms complicate the system further. Adding to the pathway are factors that contribute to the disease process and make AD worse. Among these are medications such as haloperidol and chemicals such as nicotine and beta methyl aminoalanine which are known to be biofilm dispersers. Diabetes is known to make AD worse; the likely mechanism for this is an increase in biofilms caused by hyper osmolality in the serum. Thus, making or breaking (dispersing) biofilms eventuates in more biofilms and more disease. Only a few factors ameliorate AD: low levels of vitamins D3 and K2 upregulate TLR2 causing more disease activity. Higher serum levels do the opposite. Vitamin E and L-serine are quorum sensing inhibitors; and, when present, aid in preventing biofilm formation. Last, if the microbes are killed (by penicillin) before they reach the brain or before they make biofilms, there would likely be no disease at all.
Anna Krasowska
University of Wroclaw, Poland
Title: The capric acid from Saccharomyces boulardii as an antifungal agent: a mechanism study
Time : 14:00-14:25

Biography:
Anna Krasowska is an assistant professor at the Department of Biotransformation, University of Wroclaw, Poland. Currently they are involved in the isolation and characterization of biosurfactants produced by arctic microorganisms. She has also examined the activity of lipases and proteases released into the environment by microorganisms isolated from different environments. Her research interests lie in multidrug resistance of pathogenic microorganisms like Saccharomyces cerevisiae, yeast and Candida albicans.
Abstract:
Candida albicans is a pathogenic yeast-like fungus that causes exo- and endogenous infections. C. albicans strains exhibit multidrug-resistance to commonly used antifungal agents which correlates with overexpression of Cdr and Mdr efflux pumps located in the plasma membrane. Growing resistance of pathogenic C. albicans strains to many classes of antifungal drugs has stimulated efforts to find new agents to combat more invasive infections. A selected number of probiotic organisms, Saccharomyces boulardii among them, have also been tested as potential biotherapeutic agents. S. boulardii is a yeast strain that has been shown to have applications in the prevention and treatment of intestinal infections caused by microbial pathogens. We have similarly shown that S. boulardii secretes capric acid (C10:0), which is most effective in inhibiting essential virulence factors of C. albicans, especially morphological transition, partial adhesion, as well as biofilm formation. Our latest research on the mechanism of action of capric acid and its influence on the C. albicans’ cells clearly show its interaction with the plasma membrane. Capric acid decreases fluidity, while increasing the potential of the plasma membrane. For these reasons, we have probably not observed antifungal activity of amphotericin B in the presence of capric acid. The antagonism between capric acid and amphotericin B is a strong indication for physicians to not use both compounds simultaneously in the treatment of candidiasis.
Recent Publications
1. Sellam A, Whiteway M (2016) Recent advances on Candida albicans biology and virulence F1000Res 26:2582-2590.
2. Sanguinetti M, Posteraro B, Lass-Flörl C (2015) Antifungal drug resistance among Candida species: mechanisms and clinical impact Mycoses 58:2-13
3. Kumar S, Singhi S, Chakrabarti A, Bansal A, & Jayashree M (2013) Probiotic use and prevalence of candidemia and candiduria in a PICU Pediatric Critical Care Medicine 14: e409-e415.
4. Martin I, Tonner R, Trivedi J, Miller H, Lee R, Liang X, Rotello L, Isenbergh E, Anderson J, Perl T, & Zhang X (2017) Saccharomyces boulardii probiotic-associated fungemia: questioning the safety of this preventive probiotic's use Diagnostic Microbiology and Infectious Disease 87:286–288.
5. TomiÄić Z, Zupan J, Matos T, & Raspor P (2016) Probiotic yeast Saccharomyces boulardii (nom. nud.) modulates adhesive properties of Candida glabrata Medical Mycology 54:835-845.
Tatyana Tracevska
University of Latvia, Latvia
Title: Interdisciplinary studies on Ayurvedic herbal formulation showing antibacterial effect on methicillin resistant Staphylococcus aureus
Time : 14:25-14:50

Biography:
Tatyana Tracevska is an Associate Professor of University of Latvia, Faculty of Medicine. She has her expertise in Molecular Microbiology, focusing on drug resistance mechanisms, persistent infections and epidemiological studies of pathogens such as Mycobacterium tuberculosis, Staphylococcus aureus Chlamydia trachomatis. In 2009, she received ESCMID (European Society of Clinical Microbiology and Infectious Diseases) and bioMérieux Award for Advances in Clinical Microbiology in East Central or Central Europe, and for the project entitled, luxS gene as a novel diagnostics for invasive CoNs. From 2010 to 2012, she was involved as a Head Scientist in European Social Fund co-founded project “Capacity building for interdisciplinary biosafety research” by University of Latvia. From 2016, she is a Head of effective collaboration project between University of Latvia and Arya Vaidya Pharmacy Baltics “Development of a novel herbal product for wound healing using the method of lamellar gel phase emulsion”.
Abstract:
Statement of the Problem: Emerging bacterial resistance in response to antimicrobial therapy is a world-wide problem nowadays, stimulating the search for new antimicrobial compounds. Combinatory therapy including antibacterial agents and herbal extracts, is able to combat bacterial resistance, is the promising perspective. The object of present study is a polyherbal formulation, Jathyadi Thailam, based on 13 herb infusion in coconut oil, used in Ayurvedic medicine for chronic wound (e.g. Diabetic foot ulcers) and burns healing.
Methodology & Theoretical Orientation: The antimicrobial activity of this formulation for biofilm forming and drug resistant bacteria such as methicillin resistant staphylococci, or extended spectrum beta-lactamase producing bacilli has been studied. The antibacterial efficacy of the herbal oil and its polar and nonpolar crude extracts was tested by agar dilution method and by broth microdilution method for mostly common isolates from diabetic wounds. Reference strains and clinical isolates from diabetic foot ulcers isolated in Latvian hospital were analyzed.
Findings: Interestingly, the bacteriostatic effect was shown by Jathyadi Thailam by the agar dilution method for methicillin resistant Staphylococcus aureus ATCC 38592, drug susceptible S.aureus ATCC 2848 and clinical S.aureus strain, Pseudomonas aeruginosa ATCC 2843, Enterococcus faecalis ATCC 29212 and biofilm producing S.epidermidis reference strains. No inhibition was observed for Klebsiella pnemoniae ATCC 2558, Proteus mirabilis ATCC 432351, Echerichia coli ATCC 25922 and for clinical Prowidencia spp. and ESBL producing K.pneumoniae strain. Minimal bactericidal concentration (MBC) was determined for the crude herbal polar and nonpolar extracts by the broth microdilution method. The MBC was determined as 15.6 mg/ml and 31.2 mg/ml by polar and nonpolar extracts, respectively, for MRSA ATCC 38592, 7.8 mg/ml by both extracts for S.aureus ATCC 2848, 1.95 mg/ml by both extracts for biofilm producing S.epidermidis and 62.5mg/ml by nonpolar extract for P. aeruginosa.
Conclusion & Significance: To conclude, both herbal crude extracts and the final herbal oil were found to be more effective for Gram positive bacteria, when for Gram negative. The formulation was effective against Staphyloccus aureus, the bacteria predominant in chronic diabetic wounds. Further studies will be focused on development of herbal suspension with better skin penetrating properties and aimed to inhibit the growth of Gram negative bacteria.
Recent Publications
- Dowd SE, Sun Y, Secor PR, Rhoads DD, Wolcott BM et al. (2008) Survey of bacterial diversity in chronic wounds using Pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol 8: 43.
- Dhande Priti P, Raj S, Kureshee N et al. (2012) Burn wound healing potential of Jatyadi formulations in rats. Research Journal of Pharmaceutical, Biological and Chemical Sciences. 3(4): 747-754.
- Boutoille D, Fe Ìraille A, Maulaz D et al (2008) Quality of life with diabetes-associated foot complications: comparison between lower-limb amputation and chronic foot ulceration. Foot and Ankle International, 29(11):1074–1078.
- Payne D, Gwynn M, Holmes DJ et al (2007) Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nature Reviews Drug Discovery 6(1):29– 40.
- Menon SS, Pednekar S and Singh A (2011) Wound healing efficacy of Jatyadi Taila: in vivo evaluation in rat using excision wound model, J. Ethnopharmacol. 138: 99-104.
Stef Stienstra
Scientific Advisor Royal Dutch Navy, Netherlands
Title: Cooperation in public health to fight infectious diseases in developing countries is good for the global economy
Time : 14:50-15:15

Biography:
Stef Stienstra works internationally for several medical and biotech companies as scientific advisory board member and is also an active reserve-officer of the Royal Dutch Navy in his rank as Commander (OF4). For the Dutch Armed Forces, he is CBRNe specialist with focus on (micro)biological and chemical threats and medical- and environmental functional specialist within the 1st CMI (Civil Military Interaction) Battalion of the Dutch Armed Forces. He is managing an EU CBRN CoE public health project in West Africa on behalf of Expertise France. In his civilian position, he is currently developing with MT-Derm in Berlin (Germany) a novel interdermal vaccination technology as well as a new therapy for cutaneous leishmaniasis for which he has won a Canadian ‘Grand Challenge’ grant. With Hemanua in Dublin (Ireland) he has developed an innovative blood separation unit, which is also suitable to produce convalescent plasma for Ebola Virus Disese therapy. He has finished both his studies in Medicine and in Biochemistry, Netherlands with a doctorate and has extensive practical experience in cell biology, immuno-haematology, infectous diseases, biodefense and transfusion medicine.
Abstract:
Public health systems are not always prepared for outbreaks of infectious diseases. Although in the past several public health institutes, like the French ‘Institut Pasteur’ and the Dutch ‘Tropeninstituut ‘, were prominent surveyors of infectious diseases, the investments in worldwide public health have decreased. Now more attention is given to curative healthcare compared to preventive healthcare. The recent Ebola Virus Disease outbreak in West Africa initiated a new wave of interest to invest in Worldwide Public Health to prevent outbreaks of highly contagious diseases. Zoonotic diseases are threatening as the population does not have natural nor artificial (from vaccination) immune response to new diseases like in the Ebola Virus Disease outbreak in 2014. The new strain of the Ebola Virus in West Africa was slightly less lethal, compared to other Ebola Virus strains, but the threat of spreading was far bigger as it had a longer incubation time. Most public health systems are not trained well enough to mitigate highly infectious and deadly disease outbreaks. NGO’s helping to fight the outbreak are often better trained in curative treatments and have less experience with biological (bioweapon) threats for which the military are trained for. The UNMEER mission was unique in this. It was a setting in which military and civilian actors cooperate in fighting a biological threat. Protection is essential for health workers. Smart systems must be developed to prevent further spreading of the disease, but it is not only the biosafety, which has to be considered, but also the biosecurity, as misuse of extremely dangerous strains of microorganisms cannot be excluded. Several zoonotic infectious diseases, like anthrax, smallpox and hemorrhagic fevers are listed as potential bioweapons. Therefor both biosafety and biosecurity have to be implemented in all measures to fight outbreaks of highly infectious diseases.
Recent publications
1. Moon S et. al. (2015) Will Ebola change the game? Ten essential reforms before the next pandemic. The report of the Harvard-LSHTM Independent Panel on the Global Response to Ebola. Lancet. 386(100069): 2204-21.
2. Kamradt-Scott A et. al. (2015) WHO must remain a strong global health leader post Ebola. Lancet 385(9963):111.
3. Kieny MP, Dovlo D (2015) Beyond Ebola: a new agenda for resilient health systems. Lancet 385(9963):91-92.
4. Cenciarelli O et. al. (2015) Viral bioterrorism: Learning the lesson of Ebola virus in West Africa 2013-2015. Virus Research 210: 318-326.
5. Abramowitz SA et. al. (2015) Social science intelligence in the global Ebola response. Lancet 385(9965): 330.

Biography:
Herbert B Allen specialties include dermatology and dermatopathology, skin pathology and fungal infections. He is a graduate of Johns Hopkins University School of Medicine. He has served on the boards of the American Society of Dermatology and the American College of Physicians and has published over 30 scientific articles in the fields of dermatology and dermatopathology. He is the author of ‘Keywords’ in Dermatology, a book on the language of dermatology. He's board-certified with the American Board of Dermatology and the American Board of Pathology. He is currently an Emeritus Professor in the department of Dermatology where he served as chair of the department for 14 years.
Abstract:
We recently have found biofilms in an intracellular position in Alzheimer’s disease (AD), molluscum contagiosum (MC), and psoriasis. We have noted them also in leprosy. With AD, the biofilms are made primarily by dental and Lyme spirochetes; with MC, it is the molluscum virus that “hijacks” the cell DNA and makes them; and with psoriasis, it is streptococci, primarily in the pharynx that make them. In leprosy, we have observed them inside the “globi” that are prominent in lepromatous leprosy, and they are made by Mycobacterium leprae. Thus, spirochetes, DNA viruses, gram positive bacteria, and mycobacteria can be added to the list of microbes that make intracellular biofilms. Intracellular biofilms add another layer of protection to the slime coating already in place. We have found these biofilms with routine histopathologic staining with PAS and Congo red (CR) stains. The PAS stains the extracellular polysaccharides (EPS) that are an essential ingredient in biofilms. The CR stains the amyloid that forms the infrastructure of biofilms and is another essential element. These intracellular biofilms have both been clearly shown in these diseases and exist alongside the extracellular biomasses that have previously been found. Limits to the evaluation of these findings involve the resolution power of light microscopy, selection of the tissue involved (in psoriasis, it is the tonsils, not the skin, where the biofilms are found), and the lack of a test, such as culture, to confirm the findings. The size of the organisms is important to make a biofilm, the microbes must have 10 organisms in any direction to begin the process. The resulting aggregate must then fit within the cell cytoplasm. The size ranges from 3 nM in MC to 25µM in Borrelia. These clearly are spatially able to fit within the cells. These findings are novel as to finding biofilms intracellularly in the skin and are the second time in history that a virus has been shown to make biofilms.
- Young Research Forum
Location: Redwood Suite C

Chair
Alastair Fleming
Trinity College Dublin, Ireland
Session Introduction
Nicolas Le Goff
Enzyme and Cellular Engineering Laboratory, UTC, France
Title: Recalcitrant polymers biodegradation by phylloplanic bacterial and fungal isolates: identification of efficient strains and characterization of the degradation products
Time : 17:15-17:30

Biography:
Nicolas Le Goff, after graduating as a Biological Engineer at the UTC, mostly specialized in healthcare and drug delivery, he wanted to diversify his skills and started his PhD project at the Enzymatic and Cellular Engineering (GEC) Laboratory and the Integrated Transformation of the Renewable Matte (TIMR) Laboratory of the UTC. He truly believes that biomimetism and nature can inspire research and will allow discovering and developing alternatives to actual polluting industrial processes. The approach of this work can be adapted to different environmental media to identify microbial strains of interest for a large scale of applications.
Abstract:
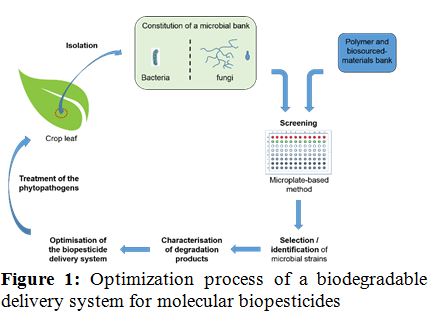
Statement of the Problem: The excessive use of chemical pesticides in agriculture represents healthcare and environmental concerns. A possible alternative to fight against phytopathogens is the use of biopesticides such as secondary metabolites or enzymes. But the stabilization of those molecules to obtain long term and higher efficiency on plant is still an issue. One proposed solution is the encapsulation of biopesticides in polymeric materials. Thus, the biodegradability of those materials by microbes present on the crops leaves has to be evaluated for the selection of the most suitable encapsulating agent. The work presented here consists in the isolation of microorganisms from crops, the screening of their degradation abilities and the identification of efficient strains.
Methodology & Theoretical Orientation: First, bacteria and fungi have been isolated from different crops leaves (corn, rapeseed, cabbage and sugar beets). Two isolation techniques were used: the suspension dilution technique and the foliar imprints method. A small scale microplate-based screening method was optimized to select both bacteria and fungi able to grow with different polymeric materials as sole carbon source. Secondly, the degradation products of the polymers were characterized with different analytical techniques (GC-MS, SEC, isotopic labeling) to identify the metabolic pathways and to prevent to impact the environment with toxic metabolites.
Findings: 117 fungal and 212 bacterial isolates have been obtained and screened for the degradation of several polymers (pHEMA, pMMA, pNIPAM, pAcrylate, pAcrylamide and bio-sourced polymer). First results show good abilities of the microbial bank to degrade mainly pHEMA and pAcrylamide.
Conclusion & Significance: Numerous microbial strains isolated in this work are able to degrade polymeric and biobased materials. This should allow the development of new biodegradable vectorization systems for the stabilization of molecular biopesticides as an alternative to chemicals, but also of bioremediation processes.
Recent Publications
- Transparency Market Research (2016) Biopesticides Market - Global Industry Analysis, Size, Share, Growth and Forecast 2015 – 2023, 95.
- Villaverde JJ (2014) Chapter 15 – Challenges of Biopesticides Under the European Regulation (EC) No. 1107/2009: An Overview of New Trends in Residue Analysis. Studies in Natural Products Chemistry 43: 437-482.
- Chilukoti N (2010) Biotechnological approaches to develop bacterial chitinases as a bioshield against fungal diseases of plants. Critical Rewiews in Biotechnology 30(3): 231-241.
- Salgado M (2015) Encapsulation of resveratrol on lecithin and b-glucans to enhance its action against Botrytis cinerea. Journal of Food Engineering 165:13–21.
- Ya-Ting X (2011) Molecularly imprinted polymer microspheres enhanced biodegradation of bisphenol A by acclimated activated sludge. Water Research 45(3): 1189-1198.
Katherine Hayes
Cork Institute of Technology, Ireland
Title: Macrolide and lincosamide resistance amongst Group B Streptococci – a growing concern?
Time : 17:30-17:45

Biography:
Kate Hayes is a Postgraduate student in the Biological Sciences at the Department of Cork Institute of Technology. Upon completion of her undergraduate degree in Biomedical Science, she was awarded a RÍSAM Scholarship by CIT. With interests in clinical microbiology and molecular biology, she is undertaking research on Group B Streptococci with a project entitled “Group B Streptococci - molecular epidemiology, pathogenic profiling and control strategies” under the joint supervision of Dr. Lesley Cotter and Dr. Fiona O’ Halloran.
Abstract:
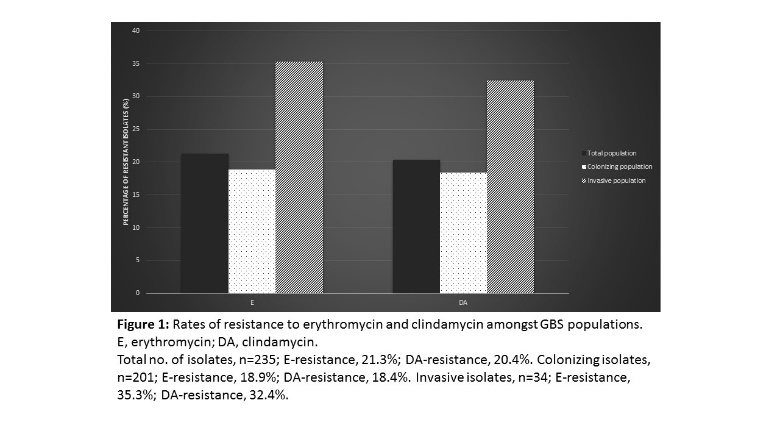
Group B Streptococcus (GBS) is the leading cause of invasive neonatal disease worldwide, with fatality rates of up to 10%. It colonizes the genitourinary tract of approximately 30% of pregnant women and in recent years it has become a growing concern amongst non-pregnant adults. Penicillin remains the first line of treatment for GBS infections, however there have been reports of reduced sensitivity in some countries. Clindamycin and erythromycin are used in cases of beta-lactam allergy, with rates of resistance to these antibiotics continuing to rise. This study aimed to investigate the incidence of antimicrobial resistance in a clinical population of GBS isolates (n=235). Antimicrobial susceptibility testing was performed according to EUCAST guidelines and the underlying mechanism of resistance was determined, both phenotypically and genotypically. Isolates were further characterized based on their serotype using molecular methods and the prevalence of the hyper-virulent ST-17 clone was investigated. Resistance to erythromycin and clindamycin was observed in 21.3% and 20.4% of the total population respectively. The c-MLSB phenotype was the most common, detected in 62% of isolates, followed by i-MLSB (20%) and M (18%) phenotypes. The rare L phenotype was also confirmed in the clinical population. ErmB was the predominant genetic determinant, identified in 84% of isolates. Both mefA/E and ermTR were each present in 18% of the isolates. Molecular serotyping analysis revealed capsular types Ia, III and II were the most common serotypes (28.1%, 24.7% and 14% respectively). Evidence of capsular switching was observed in one GBS isolate, which has implications for vaccines that target the capsular polysaccharide. This work highlights emerging trends in antimicrobial resistance amongst the Irish GBS population and emphasizes the need for continued monitoring of antibiotic resistance and serotype distribution in the population.
Recent Publications
1. Hawkins PA, et al. (2017) Cross-resistance to lincosamides, streptogramins A and pleuromutilins in Streptococcus agalactiae isolates from the USA. Journal of Antimicrobial Chemotherapy 5: 4544.
2. Arana DM, et al. (2014) First clinical isolate in Europe of clindamycin-resistant group B Streptococcus mediated by the lnu(B) gene. Revista española de quimioterapia: publicacioÌn oficial de la Sociedad Española de Quimioterapia 27: 106–9.
3. Lamagni TL, et al. (2013) Emerging trends in the epidemiology of invasive group B streptococcal disease in England and Wales, 1991-2010. Clinical infectious diseases 57: 682–688.
4. Malbruny B, et al. (2011) Cross-resistance to lincosamides, streptogramins A, and pleuromutilins due to the lsa(C) gene in Streptococcus agalactiae UCN70. Antimicrobial Agents and Chemotherapy 55: 1470–4.
5. Kimura K, et al. (2008) First Molecular Characterization of Group B Streptococci with Reduced Penicillin Susceptibility. Antimicrobial Agents and Chemotherapy 52: 2890–2897.
Albertus Geurkink
University of Groningen, Netherlands
Title: Transcriptomics as a tool to monitor and predict the behaviour of the microbiome in an industrial salt waste water treatment plant
Time : 17:45-18:00

Biography:
A K Geurkink works in the laboratory of G J W Euverink, at the Faculty of Science and Engineering, University of Groningen. He is also a part of Bioclear b.v, The Netherlands. His research interests are in Applied Microbiology, Biotechnology and Water Microbiology/bio-technology.
Abstract:
In waste water treatment plants, polluted water is purified with the help of complex ecosystems of many different bacteria. In these systems, many different desirable but also undesirable microbial activities play a role. To achieve detailed insight in the microbial activities of these processes in a waste water treatment plant, metagenomic tools can be applied. Until now, transcriptomics tools are almost exclusively reserved for applications in fundamental research. One of the reasons is that metagenomic techniques are complex and generates a very large dataset. Furthermore, in most cases it takes too long to obtain practical answers and solutions to questions raised in the ‘real world’. The costs associated with such an approach are also very high compared to the costs of measuring a set of standard physical and chemical parameters. However, transcriptomics results in an enormous amount of knowledge about the microbial processes in a waste water treatment plant that can be used to improve the quality of the effluent. Therefore, new pragmatic ways to bridge the gap between transcriptomics and business are required. This approach is characterized by the strong cooperation between fundamental research and a practical application. It distinguishes itself by combining Next Generation Sequence (NGS) data of DNA and RNA extracted from a waste water treatment plant at specific time points. NGS data will give detailed insight into the population and activities of a sample while (high throughput) quantitative-PCR is used to get insight into the dynamics of selected set of target genes. Mathematical analysis of the historical data combined with the chemical/physical characteristics of the influent water allows us to predict and control the microbial behavior and thereby the performance of the waste water treatment plant.
Claire Baranger
Université de Technologie de Compiègne, France
Title: “Myco-fluidics†for soil remediation: developing a microfluidic tool to monitor the solubilization and uptake of a persistant soil pollutant in filamentous fungi

Biography:
Claire Baranger graduated from the Ecole Supérieure de Biotechnologie de Strasbourg, France, in 2015. She is a trained biotechnologist with experience in microbiology and enzymology, and has a specific interest in mycology and environmental biotechnologies. She is currently pursuing a PhD at the Université de Technologie de Compiègne in France.
Abstract:
Bioremediation appears as a relatively low-cost solution for the restoration of soils contaminated with persistent organic pollutants. Among these pollutants, high molecular weight polycyclic aromatic hydrocarbons (PAHs) display a high resistance to degradation and low bioavailability.
Filamentous fungi are known for their ability to degrade complex substrates, and some soil fungi, such as Talaromyces helicus, showed promising results regarding the biodegradation of PAHs. However, the mechanisms involved in the incorporation and biodisponibility modifications of these pollutants remain poorly understood.
In this context, the Myco-fluidics project aims at using microfluidics to mimic the soil microenvironment and monitor the solubilization and uptake of pollutants in filamentous fungi in vivo. This approach allows direct microscopic observations at a cellular scale, in complement with a characterization of the degradation in macroscopic cultures.
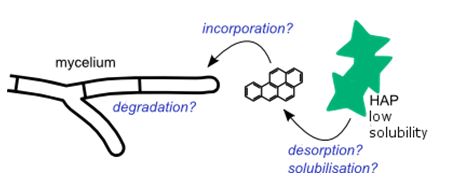
Preliminary results show the intracellular storage of benzo[a]pyrene in T. helicus hyphae, and suggest the release of tensio-active compounds likely to promote the desorption and stabilization of HAPs in the aqueous phase. In a broader perspective, this soil-on-a-chip model could be used as a new tool to investigate parameters affecting the biodegradation of pollutants, in order to develop more efficient fungus-mediated soil cleanup strategies.
Recent Publications
- Bhardwaj, G. (2013). Biosurfactants from Fungi: A Review. J. Pet. Environ. Biotechnol. 4
- Fayeulle, A., Veignie, E., Slomianny, C., Dewailly, E., Munch, J.-C., and Rafin, C. (2014). Energy-dependent uptake of benzo[a]pyrene and its cytoskeleton-dependent intracellular transport by the telluric fungus Fusarium solani. Environ. Sci. Pollut. Res. 21, 3515–3523.
- Held, M., Edwards, C., and Nicolau, D.V. (2011). Probing the growth dynamics of Neurospora crassa with microfluidic structures. Fungal Biol. 115, 493–505.
- Marco-Urrea, E., García-Romera, I., and Aranda, E. (2015). Potential of non-ligninolytic fungi in bioremediation of chlorinated and polycyclic aromatic hydrocarbons. New Biotechnol. 32, 620–628.
- Stanley, C.E., Grossmann, G., Casadevall i Solvas, X., and deMello, A.J. (2016). Soil-on-a-Chip: microfluidic platforms for environmental organismal studies. Lab Chip 16, 228–241.
- Special Talk
Location: Redwood Suite C
Chair
TBD
TBD
Session Introduction
Alain Richard
National Research Council of Canada (NRC), Québec, Canada
Title: Tips for a Successful Technology Transfer in Life Sciences

Biography:
Alain Richard completed his BSc in Microbiology and MSc in Microbiology-Immunology & PhD). He has dedicated his 20 years’ career to innovation in biotechnology and biopharma, acting at crossroads between science and business: 8 years in biotech companies and more than 12 years in technology transfer in the Province of Québec, Canada (university or government - industry liaison). He is currently active in this particular field at the National Research Council of Canada as a Client Relationship Leader in Human Health Therapeutics.
Abstract:
The relevance of technology transfer has been demonstrated from the early stages of microbiology (e.g. industrial cases with Louis Pasteur). Technology transfer allows the deployment of science into society. It is an evolving field of knowledge combining science and business approaches. It is a process that requires focus and persistence. There are some essential parameters to consider in that process. There are intellectual property aspects such as confidentiality, patenting, individual and institutional commercial rights. The developmental stage of a technology is key to deciding what steps to take to increase the chances of success of the transfer. It means partnering with the appropriate organizations involved along the process. There are many mechanisms to add value to an invention disclosure to make it more amenable for a transfer in the forms of a license or a start-up.
Some typical agreements are commonly used in technology transfer, such as confidentiality agreements, material transfer agreements, R&D licenses with or without commercial option, commercial license agreements, and creation of spin-off companies. Excellent science does not necessarily mean an entry to the market. Understanding the drivers and the needs of a relevant market can prove to be difficult, but there are some ways to overcome this. There is a cultural encounter between business and science; both fields of knowledge have to reach common goals. The conditions sought in pharmaceutical sector for in-licensing are different than in other fields of life sciences due to the multi-million-dollar cost of clinical research trials prior to market authorization. There are common indicators used in technology transfers, from rules of thumb to complex financial valuations. This presentation aims to provide some insight on the elements described above.
- Industrial Microbiology, Food Microbiology, Vaccines and Anti-Microbials, Microbial Immunology, Molecular and Medical Microbiology
Location: Redwood Suite C

Chair
Sarah Louise Cosby
Queen’s University Belfast & Agri-Food & Biosciences Institute, UK

Co-Chair
Pietro Mastroeni
University of Cambridge United Kingdom
Session Introduction
Pavel Dibrov
University of Manitoba, Canada
Title: A novel antibiotic effectively inhibiting an unconventional target, Na+-translocating NADH:ubiquinone oxidoreductase
Time : 12:25-12:50

Biography:
Abstract:
Statement of the Problem: The present crisis in antibiotic development and administering was precipitated not only by decades of global misuse of broad-spectrum antibiotics and resulting proliferation of multi-drug resistant strains, but also by a general strategy in antibiotic design, when a very limited set of prokaryotic enzymes and metabolic pathways was ever targeted. The situation calls for urgent intensification of the search for non-traditional antimicrobial targets. Methodology & Theoretical Orientation: The Na+-translocating NADH:ubiquinone oxidoreductase (Na+-NQR) is a key respiratory enzyme in many aerobic pathogens, including a widespread and notoriously difficult to treat Chlamydia trachomatis. Inhibition of Na+-NQR was predicted to arrest bacterial energization and proliferation, and ultimately disrupt the infectious process. To examine this prediction, a new furanone inhibitor of Na+-NQR, PEG-2S, was designed and assayed for its anti-chlamydial activity in cell culture models of infection. Findings: The presented work confirmes that Na+-NQR is critical for the Chlamydia trachomatis infectious process. A newly designed PEG-2S inhibited intracellular proliferation of Chl. trachomatis with a half-minimal concentration in the submicromolar range without affecting the viability of mammalian cells or selected species representing benign intestinal microflora. Infection by Chl. trachomatis increased H+ and Na+ concentration in the infected mammalian cell. Addition of PEG-2S blocked these changes in ion balance induced by Chl. trachomatis infection. PEG-2S also inhibited the Na+-NQR activity in sub-bacterial membrane vesicles isolated from Vibrio cholerae when added at very low (nanomolar) concentrations. Conclusion & Significance: The obtained results demonstrate that Na+-NQR is critical for the bacterial infectious process and is susceptible to a precisely targeted bactericidal compound in situ. PEG-2S opens a new line of narrowly targeted inhibitors of NQR and is serving as a molecular platform for the development of “individually tailored” anti-NQR remedies narrowly targeting specific pathogens.
Boris V Nikonenko
Central Institute for Tuberculosis, Russia
Title: Activity of new indole derivatives against Mycobacterium tuberculosis and Mycobacterium avium in HIV combined mycobacteriosis
Time : 14:00-14:25

Biography:
Boris V Nikonenko MD, Ph.D., Doctor of Medical Sciences is a leading Researcher at Central Research Institute for Tuberculosis in Moscow. He is an expert in immunology and genetics of experimental tuberculosis and other mycobacterial infections, as well as testing of TB vaccines and drugs. Along with colleagues, he mapped three loci in the mouse genome that regulates the course of TB infection and immunity and revealed the role of other (known) genes in mouse ant-TB resistance and immunity. For 12 years, he worked in biopharm company Sequella, Inc. (Rockville, USA) testing the activity of established drugs against M. tuberculosis, M. avium, M. abscessus, Candida in the mouse models.
Abstract:
Given the data of high-level activities of original lead compounds, obtained earlier at All-Union Scientific Research Chemical-Pharmaceutical Institute (does not exist now) and A N Nesmeyanov Institute of Organoelement Compounds, Russian Academy of Sciences (INEOS RAS), we designed now about 300 multi-target compounds in which electron donor (indole and and ferrocene) and electron acceptor pyridine scaffolds, including INH, are linked via N-N-containing pharmacophore groups, either linear or within the heterocyclic structure. Compounds were tested at the Central Institute for Tuberculosis (CNIIT). Results demonstrated that about 40 agents exhibited proven high activity in vitro and ex vivo against both drug-sensitive M. tuberculosis Ð37Rv (MIC = 0.05-2 μg/mL) and the isoniazid (INH)-resistant clinical isolates of M. tuberculosis CN-40 (MIC = 0.018-4.44 μg/mL), as well as against M. avium (MIC = 0.05-1.5 μg/mL for 15 compounds) and other non-tuberculosis HIV co-infections. Thus, these agents were virtually as active as INH against M. tuberculosis H37Rv; however, unlike INH, those showed remarkable activity against the INH-resistant M. tuberculosis strain and M. avium. 3-triazeneindole TU112 demonstrated high level activity against dormant non-culturable M. tuberculosis. In addition, about 80 compounds among the tested ones show MIC values in the range of ≥2 and ≤ 10 μg/mL, thus ~40% of the compounds exhibited appreciable anti-TB activity. The compounds are hybrid molecules designed on a basis of privileged scaffolds – 3-indolyl (substituted at positions 1,2,5) and 2-4-pyridines including isoniazid one. The strategy of designing new agents based on a concept of molecular hybridization, as well as a recently emerging concept of development membrane-active (redox-mediated) agents as a strategy for treating persistent infections.
Recent Publications
1. Nikonenko B, Reddy VM, Bogatcheva E, Protopopova M, Einck L, Nacy CA (2014) Therapeutic efficacy of SQ641-NE against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 58(1): 587-9.
2. Kondratieva T, Azhikina T, Nikonenko B, Kaprelyants A, Apt A (2014) Latent tuberculosis infection: what we know about its genetic control? Tuberculosis (Edinb). 94(5): 462-8.
3. Velezheva V, Brennan P, Ivanov P, Kornienko A, Lyubimov S, Kazarian K, Nikonenko B, Majorov K, Apt A (2016) Synthesis and antituberculosis activity of indole-pyridine derived hydrazides, hydrazide-hydrazones, and thiosemicarbazones. Bioorg Med Chem Lett. 26(3): 978-85.
4. Moore JH, van Opstal E, Kolling GL, Shin JH, Bogatcheva E, Nikonenko B, Einck L, Phipps AJ, Guerrant RL, Protopopova M, Warren CA (2016) Treatment of Clostridium difficile infection using SQ641, a capuramycin analogue, increases post-treatment survival and improves clinical measures of disease in a murine model. J Antimicrob Chemother. 71(5): 1300-6.
5. Nikonenko BV, Kornienko A, Majorov K, Ivanov P, Kondratieva T, Korotetskaya M, Apt AS, Salina E, Velezheva V (2016) In vitro activity of 3-triazenoindoles against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 60(10):6422-4.
Pierre-Yves Morvan
Codif Technologie Naturelle, France
Title: Effects of stressful life on human skin biota

Biography:
Abstract:
Marcin Åukaszewicz
University of Wroclaw, Poland
Title: Microbial production of biopolymers using solid-state fermentation with Bacillus subtilis natto
Time : 16:45-17:10

Biography:
Marcin Åukaszewicz is a Polish biologist, with his research interest and specialization in the fields of molecular biology, biotechnology and microbiology. He did his PhD in Catholic University, Belgium and later became became an assistant professor at the Institute of Genetics and Microbiology, University of WrocÅ‚aw. He is currently the dean of the Department of Biotransformation, at the Faculty of Biotechnology, University of WrocÅ‚aw.
Abstract:
Biopolymers are obtained from a wide range of microorganisms and plants. Microbial polymers are produced by the fermentation process or by chemical polymerisation of monomers. They have a wide variety of applications and can potentially replace synthetic materials. Using industrial by-products to produce exopolysaccharides can reduce the costs associated with their biosynthesis and can solve the problem of waste management after their production. It is important to select an appropriate microorganism that does not endanger human health. Solid-state fermentation (SSF) has biotechnological advantages such as higher fermentation productivity and higher product concentration. Over past five years, there have been significant developments in this method, unfortunately these advances have mainly occurred at the laboratory scale. Bacillus subtilis natto is a well-known GRAS microorganism that produces the biopolymer levan. Levan is a polymer consisting of D-fructofuranose units joined by β-(2,6) linkages and is produced by both plants and bacteria. Microbial levan potentially has a very wide range of applications in the food industry, cosmetics, pharmaceuticals, and medicine. Industrial application of levan depends on its molecular weight, which can be modulated using different strains, as well as culture conditions such as temperature, pH, agitation, carbon source, and other medium components. The aim of this research was to obtain levan in SSF using rapeseed meal as the main substrate and different Bacillus subtilis natto strains. A new bioreactor for SSF was designed. Suitable methods for product separation and purification were also investigated. The purification process was performed with low-pressure liquid chromatography, using different types of column fillers. The molecular weight distribution of levan produced in the SSF process was determined using gel permeation chromatography, and 1H NMR (Proton nuclear magnetic resonance) was used to identify levan. We observed some differences in the molecular weight range of levan obtained from the cultivation of Bacillus strains in submerged fermentation and SSF. Various molecular weight levans may have different applications due to their specific properties.
Recent Publications
1. Esawy M A, Abdel-Fattah A M, Ali M M, Helmy W A, Salama B M, Taie H A A, Hasheman AM, Awad G E A (2013) Levansucrase optimization using solid state fermentation and levan biological activities studies. Carbohydrate Polymers 96: 332–341.
2. Kreyenschulte D, Krull R, Margaritis A (2012) Recent advances in microbial biopolymer production and purification. Critical Reviews in Biotechnology 34: 1–15.
3. Porras-Domínguez JR, Ávila-Fernández A, Miranda-Molina A, Rodríguez-Alegría ME, López Munguía A (2015) Bacillus subtilis 168 levansucrase (SacB) activity affects average levan molecular weight. Carbohydrate Polymers 132: 338–344.
4. Öner ET, Hernández L, Combie J (2016) Review of Levan polysaccharide: from a century of past experiences to future prospects. Biotechnology Advances 34: 827–844.
5. Thomas L, Larroche C, Pandey A (2013) Current developments in solid-state fermentation. Biochemical Engineering Journal 81: 146–161.
Petra Dersch
Helmholtz Centre for Infection Research, Germany
Title: Role of Type III secretion system and the CNFY toxin for the virulence of enteric Yersiniae

Biography:
Abstract:
Herbert B. Allen
Drexel University College of Medicine, USA
Title: Leprosy from a Different Perspective

Biography:
Herbert B Allen specialties include dermatology and dermatopathology, skin pathology and fungal infections. He is a graduate of Johns Hopkins University School of Medicine. He has served on the boards of the American Society of Dermatology and the American College of Physicians and has published over 30 scientific articles in the fields of dermatology and dermatopathology. He is the author of ‘Keywords’ in Dermatology, a book on the language of dermatology. He's board-certified with the American Board of Dermatology and the American Board of Pathology. He is currently an Emeritus Professor in the Department of Dermatology where he served as chair of the department for 14 years.
Abstract:
We have recently written about leprosy and its treatment that has proven to be an incredible achievement in medical history. This biblical disease has been brought under control and the worldwide incidence has decreased from 12,000,000 in 1975 to 800,000 in 2015. The only thing that changed in that time span was the addition of rifampin to the treatment regimen. Rifampin “pokes holes” in biofilms and allows dapsone to penetrate and kill the mycobacteria inside. Prior to rifampin, the organism was becoming resistant to dapsone. Clofazamine was also added, but it was useful mainly in relation to managing the reaction states in the disease, mostly erythema nodosum leprosum. Even though rifampin was added as another antibiotic because of resistance, its action was too short lived (3-5 hours half-life) for its activity given the prescribed dosage of once per month. Also, resistance develops rapidly with rifampin, so it seems evident it is behaving differently from its activity as an antibiotic. That different activity is biofilm dispersion. Biofilm is most assuredly present in leprosy because we have observed it not only in “globi” in the skin, but also in the liver, spleen, and kidneys as “secondary amyloidosis of chronic disease.” It stains histopathologically with PAS which stains the polysaccharides make up the bulk of the biomass and with Congo red that stains the amyloid that makes up biofilm infrastructure. TLR2 has been shown to be activated by biofilms, and TLR2 itself has been shown to activate IL4, IL 10 both of which are present in lepromatous leprosy. The fortuitous choice to include rifampin in the multidrug regimen reversed the course of leprosy and offers hope that other chronic diseases would behave similarly.
- Workshop
Location: Redwood Suite C
Session Introduction
Joachim Wink
Helmholtz Centre for Infection Research, Germany
Title: Polyphasic Taxonomy – The Class Actinobacteria

Biography:
Joachim Wink has completed his PhD in 1985 from Frankfurt University. He then went to the pharmaceutical industry and started his career at the Hoechst AG, where he was responsible for the strain collection and specialized in the cultivation and taxonomic characterization of Actinobacteria and Myxobacteria. During the years, he was responsible for the strain library within the pharmaceutical research and a number of screening projects with Hoechst Marion Russel, Aventis and Sanofi. In the year 2005, he did his habilitation at the Carolo Wilhelma University of Braunschweig and in 2012, he went to the Helmholtz Centre for Infection Research in Braunschweig, where he founded the working group of the strain collection with its focus on Myxobacteria. Here, he is now working on the isolation and taxonomic characterization of Myxobacteria and Actinobacteria, as well as, the analysis of their secondary metabolites. He has published more than 50 papers on secondary metabolites and the taxonomy of the producing microorganisms in reputed journals, a number of reviews, as well as, book chapters and more than 35 patents. He is the Editorial Board Member of a number of international journals.
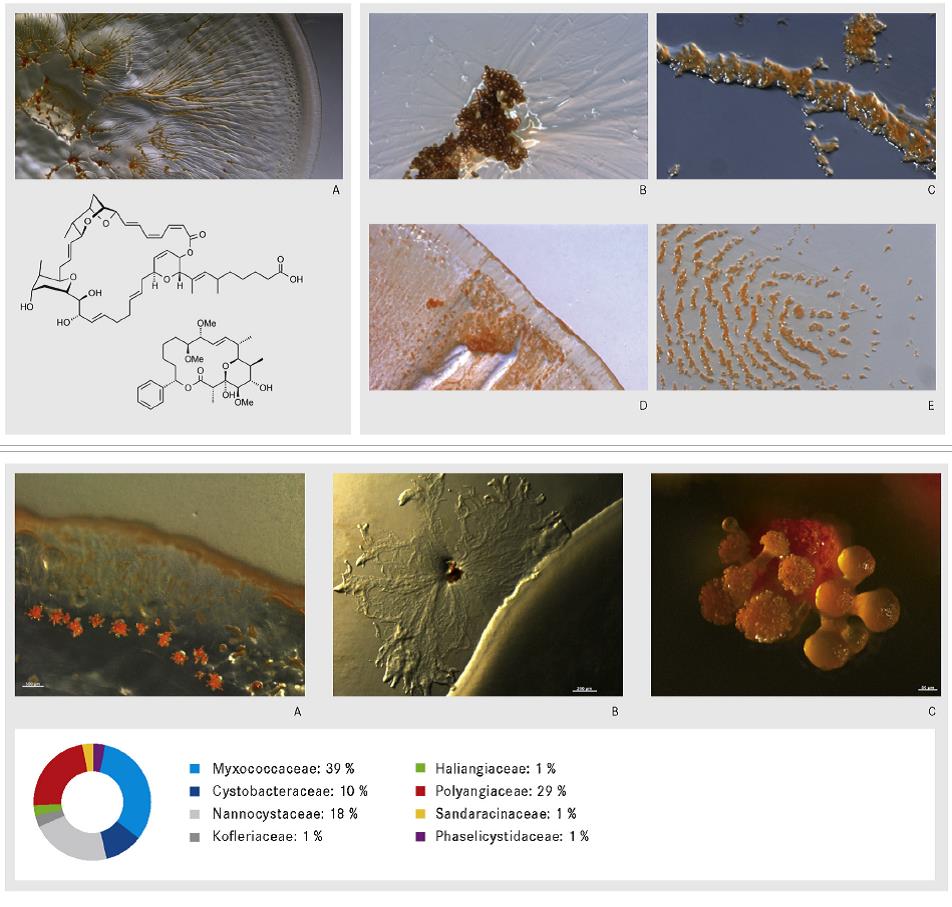
Abstract:
The myxobacteria with the order Myxococcales (TCHAN, POCHON and PRÉVOT 1948) belong to the Gram negative Proteobacteria and have first been described in detail by Thaxter in 1892 in the Botanical Gazette. Most known myxobacteria occur in soil and frequently develop on decomposing plant material, the bark of living trees or animal dung. Both in nature and in the laboratory their presence may be detected through the appearance of fruiting bodies. Since the introduction of Epotilon as anticancer therapeutic on the market, the myxobacteria have their place in the group of industrial important bacteria. The biology of myxobacteria is mainly characterized by two features that are gliding on surfaces without any locomotion organelles and the formation of fruiting bodies. Myxobacteria also have a high GC contend and very huge genomes with a size of about 10 Mb which correlates with their ability to produce many different secondary metabolites. Until today, the taxonomy of the myxobacteria is basing on the morphological features together with the 16S rRNA sequence, in addition, we have established a number of chemotaxonomic and molecular biological markers, which can be used in myxobacterial taxonomy. A short overview on the history of myxobacteria, the fruiting body formation, its taxonomic classification and additional methods for characterization, as well as, their potential to produce bioactive compounds is given in this talk.